22 December 2017
Scientists engineer microbes to form ‘memories’ of their environment
Posted by Lauren Lipuma
Inserting chemically sensitive genes into the DNA of bacteria can produce lasting “memories” of their environment and show scientists how they communicate.
Credit: Pixabay.
By Jeremy Rehm
Microbes like bacteria aren’t conscious enough to form memories, but a group of scientists in Texas developed a new way for them to do so at the genetic level.
Researchers report they’ve successfully engineered microbes to report on their environments and form genetic “memories” of the event. It’s a tool that could help scientists better understand chemical cycling on Earth and how microbes share information like antibiotic resistance with one another, according to the researchers.
“We’re hoping this will help foster collaboration between synthetic biologists and microbiologists,” said Emily Fulk, a graduate student at Rice University in Texas and who presented her preliminary work at the 2017 American Geophysical Union Fall Meeting in New Orleans. “I’m just really excited to actually start applying this.”
Fulk works with soil bacteria, microbes that serve an important role in decomposing dead organisms and “fixing” nitrogen from the atmosphere into forms that plants and animals can use. Many questions remain unanswered about these bacteria, including how they respond to environmental conditions like drought or excessive fertilizer. Answering those questions requires long periods and constant monitoring that are impractical using current methods, so Fulk turned to synthetic biology.
Biologists have previously modified genes within a microbe’s plasmid, its circular chromosome of DNA, by inserting a specific gene that activates when in the presence of a chemical of interest. This way, the microbe produces a signal in response to an environmental stimulus.

Schematic showing how microbes can detect chemicals in their environment using a gene promoter region (sensor) to activate the gene reporter (mht) and eventually produce a gas signal (CH4Br: methyl bromide).
Credit: Emily Fulk.
Researchers interested in detecting environmental nitrates, molecules found in fertilizers, for example, could insert a nitrate-sensitive gene into the microbe’s plasmid. Then, when the microbe senses nitrates in its surroundings, it activates the gene to produce a signal or “report” to the scientists.
“You can think of it like a light switch and a lightbulb, where the sensor is like the light switch and signal the lightbulb,” Fulk said. “You can look at the lightbulb, or signal produced, and say, ‘OK, if we see that the ‘light’s on, we know that the switch has been triggered.’”
Traditionally, the light analogy was literal: microbes were designed to glow or fluoresce when they detected a specific chemical. But in soil, seeing fluorescence proves challenging. Instead, Fulk and her colleagues developed microbes that report by producing a gas.
That still left an outstanding challenge, though. To produce a signal, microbes must be alive, and because most microbes live only on the order of several hours, their report is short-lived.
Fulk resolved the issue by taking genetically engineered microbes one step further. She designed a microbe that could not only produce a gas in response to a chemical signal but also remember it had produced the gas signal long after the activating chemical disappeared and microbe died. In other words, she designed it to have a memory.
“Using memory, we can now say, ‘Let’s incubate our microbes for a week, a month, or a season,’ and then at the end of it look at the microbes and see what they’ve seen over the entire period rather than only at a specific time point,” Fulk said.
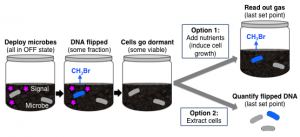
Schematic depicting how microbe “memory” (flipping the DNA) can later be uncovered through either by reactivation with nutrients or by extracting the DNA.
Credit: Emily Fulk.
In a clever design, Fulk inserted something never done before: separate “light switch” sensor and “lightbulb” reporter genes. In her setup, the reporter gene must be modified before it can produce its gas signal. The light switch gene, activated when the microbe eats the sugar arabinose, codes for an enzyme capable of modifying the reporter gene. Once the light switch gene is activated, it modifies and turns on the reporter gene, and the microbe produces a gas.
The modification to the reporter gene can’t be reversed, however, so the microbe keeps a genetic ‘memory’ of the detection. Scientists can later search for this genetic change shortly after the microbe was exposed to the chemical or long after the microbe died.
“I get blown away by this every day,” Fulk said. “We’re really excited that we have the nuts and bolts worked out.”
The new research heralds what Fulk expects will be a long line of future applications. Inserting detectors sensitive to environmentally relevant chemicals can answer long-standing questions for soil and microbial ecologists, she said.
“The idea is if we can make this a plug-and-play Lego-type system, where you pick your reporter, your ‘memory,’ your desired chemical to sense, and your microbial host, then it can be customized to help address any questions you want,” Fulk said.
“You can sometimes detect undetectable materials with these microbes,” said Caroline Masiello, the principal investigator of the research group at Rice University. Researchers could use the method to study poorly-understood aspects of the sulfur cycle or bacterial methane production, she said. But while exciting, the direct application in the environment seems unlikely.
For now, Fulk and her colleagues are working with environmental engineers to study how antibiotic resistance transfers between microbes in waste water treatment centers and in soil environments. By building a donor microbe with a light switch gene and a receiver microbe with the lightbulb gene, they could determine when the genes come together to produce a “memory.” And knowing that, in turn, will help them identify microbe “spies” they could use in the future, Fulk said.
Jeremy Rehm is a science communication graduate student at UC Santa Cruz. See more of his work at www.jeremyrehm.com or follow him on Twitter @jrehm_sci.


 GeoSpace is a blog on Earth and space science, managed by AGU’s Public Information staff. The blog features posts by AGU writers and guest contributors on all sorts of relevant science topics, but with a focus on new research and geo and space sciences-related stories that are currently in the news.
GeoSpace is a blog on Earth and space science, managed by AGU’s Public Information staff. The blog features posts by AGU writers and guest contributors on all sorts of relevant science topics, but with a focus on new research and geo and space sciences-related stories that are currently in the news.